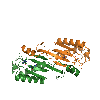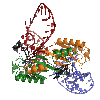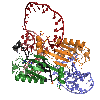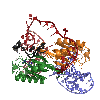
The research in the laboratory is aimed at providing a mechanistic understanding of the control of the biological activity of macromolecular complexes and assemblies using structural approaches such as X-ray crystallography and cryo-electron microscopy as a principal tool. In particular, our research focuses on understanding the genome rearrangements that occur when discrete segments of DNA are moved around within cells. For example, DNA transposons reside in host cell genomes and can be moved (or copied) into new locations by self-encoded enzymes, and we are studying how these enzymes function. DNA can also originate from outside the cell - as in the case of DNA viruses - and we would like to understand how integrating viruses infect cells.
Among the simplest transposition systems are those of prokaryotic insertion sequences (IS). We have been investigating one particular IS, ISHp608, originally isolated from the pathogenic bacterium, Helicobacter pylori. The transposase which moves ISHp608, known as TnpA, has only 155 amino acids and is among the smallest of all known transposases. We used a combined structural and biochemical approach to show that this transposase uses a completely novel mechanism for moving DNA which involves only single-stranded DNA. Furthermore, these structures revealed that TnpA recognizes both the ends of the mobile element and its integration target site through DNA-DNA interactions rather than using a site-specific DNA binding domain: target site recognition is accomplished by base pairing interactions between the target site and an internal segment of transposon DNA.
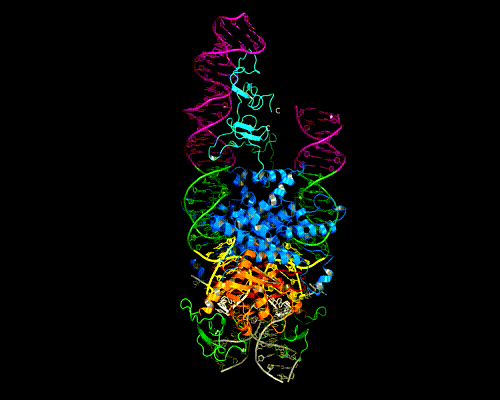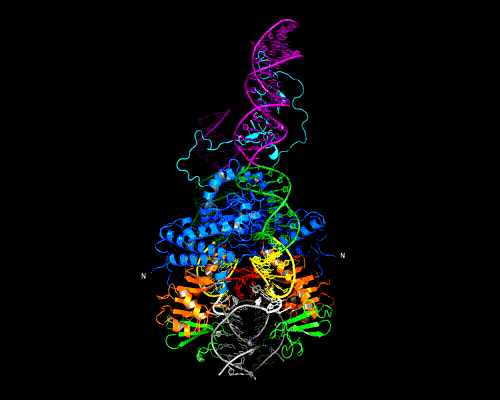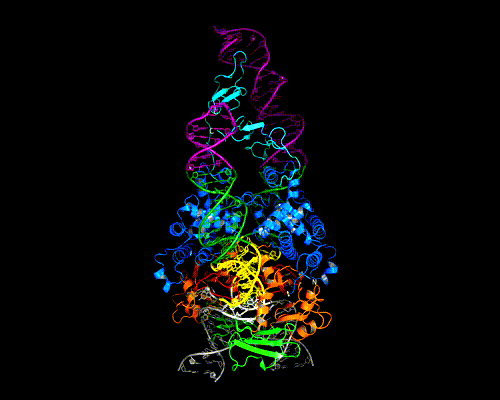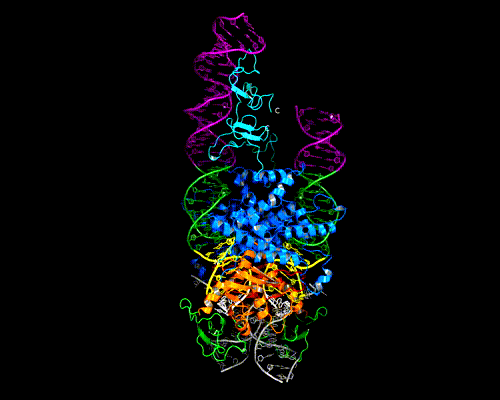
In bacteria, the movement of DNA by transposition is a crucial aspect of bacterial adaptation to new environments. For example, many bacterial transposons encode antibiotic resistance proteins in addition to those needed for their own mobility. We are interested in investigating the link between the emergence of antibiotic resistance and mobile genetic elements at the mechanistic level. Our current focus is on those transposons that move using a copy-out-paste-in mechanism, and we have recently solved by cryo-EM several structures corresponding to steps along this pathway. For the transposon we are studying, ISCth4, the structures revealed an unusual asymmetric dimeric transpososome in which an array of N-terminal domains from one monomer bind tightly to a single transposon end and the catalytic domains move to accommodate and direct the subsequent reactions.
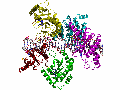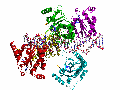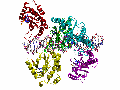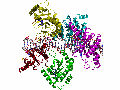
The adeno-associated virus (AAV) is a small parvovirus that is able to integrate its genome site-specifically into human chromosome 19. This process appears unique to AAV, and is important because site-specific integration is a highly desirable property of vector systems used in gene therapy. To date, there are no site-specific systems available for clinical use. We believe that understanding how the AAV integration system works could provide ideas leading to the design of clinically useful systems. Our research focuses on the structural biochemistry of the AAV replication initiator protein, called Rep, that is not only needed for replication but also for site-specific integration. We have solved several structures of the site-specific DNA binding domain of Rep and its complexes with DNA. These have led us to propose a model for Rep assembly on DNA and the initiation of viral replication, which we continue to investigate in the context of both the full-length protein and the helicase domain.
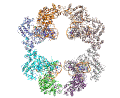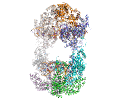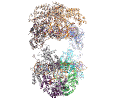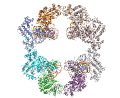
A main focus in the laboratory is the mechanisms of eukaryotic DNA transposition. Transposition has been a fundamental shaping force during evolution. For instance, it has been established that over 50% of the human genome is transposon-derived. Recent efforts have been centered on the Hermes transposase from the insect species Musca domestica. Hermes is a member of one of the largest classes of eukaryotic transposons, the hAT transposons, which has representatives scattered among the genomes of insects, plants, and even humans. We have solved the three-dimensional structure of Hermes complexed with a segment of transposon end DNA which revealed an unexpected surprise: before DNA binding, Hermes is organized as a large ring of eight subunits, a very unusual arrangement for a DNA transposase. It appears that this octameric assembly is important so that Hermes can recognize its transposon ends which are several hundred basepairs long. More recently, we reported three X-ray structures of the Hermes transposase in complex with DNA that represent steps along the flanking hairpin-forming process. Our current studies with Hermes are focused on the question of why its transposition requires such long transposon ends.
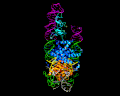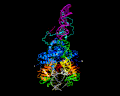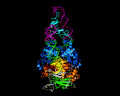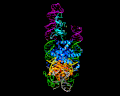
PiggyBac, an active moth transposon, is a widely used transposon in genomic applications with a number of distinctive properties. Unlike other transposons, at the excision step it liberates itself by forming hairpins on its transposon ends while leaving behind precisely complementary single-stranded overhangs which can be repaired without leaving excision footprints. This property of "seamless excision" is very valuable in applications involving genome modifications to human cells. Also distinctive is that it integrates into new sites with a strict requirement for an TTAA target sequence. We have been probing the mechanistic basis of these excision and targeting properties, and recently solved the cryo-EM structures of the piggyBac transposase bound to its hairpin end and also at the step immediately following integration into a TTAA target. These structures provide the first insight into how the unusual transposition properties of piggyBac are structurally linked.
Another superfamily of eukaryotic DNA transposons that intrigues us are the Helitrons. Although no currently mobile Helitrons have been identified, they must once have been very active, as their remnants are widespread throughout the eukaryotic kingdom. Unlike other known eukaryotic DNA transposons, Helitron insertions in the host genome are not bordered by target site duplications, suggesting a replicative transposition mechanism that differs substantially from the cut-and-paste mode of transposition used by all other currently characterized eukaryotic systems. We have been investigating a reconstituted Helitron transposon, Helraiser, which is active in cells, and are building on in vivo assays and in vitro biochemical studies to work towards structural information.

Casposons have been proposed to be novel mobile elements that are the precursor of bacterial and archaeal adaptive immune systems. They contain genes encoding Cas1 homologs that we have shown are active DNA integrases, and we recently solved the X-ray crystal structure of a complex between a casposase, its target site, and its casposon ends. It seems that the casposase is remarkably eclectic in the types of substrates it will integrate but does so site-specifically. Our structural results led us to propose an evolutionary process in which an ancestral tetrameric casposase became the CRISPR-Cas integrase that is central to spacer integration.
The Transposition Pathway of IS608
This movie presents our model of the transposition mechanism of IS608. We have recently solved the structures of the IS608 transposase (TnpA) bound to six different oligonucleotides and have captured sequential states along the transposition pathway. It's rather complicated, so we have tried to help by coloring the left end of the transposon in RED, the right end in BLUE, and the donor DNA that flanks the transposon in BLACK. The monomers of the TnpA dimer are shown in green and orange.
The movie begins with the TnpA dimer alone. As TnpA recognizes and binds short hairpins located close to the transposon ends, there is a reorganization of the active site. This entails the movement of an alpha helix across the face of the beta sheet. The helix contains the catalytic tyrosine residue (shown here in green and orange ball-and-stick), and its movement results in the correct assembly of the active site, creating a binding site for the metal ion cofactor (the little pink ball).
In the next step, TnpA cleaves both transposon ends which are present in single-stranded form. When it does this, it becomes covalently attached to the 5' end at the break. This means that at the left end, TnpA is attached to transposon DNA but at the right end, TnpA is attached to the flanking DNA.The next step requires a little imagination since we haven't crystallized this part of the reaction yet: The biochemically observed products are produced if the helices containing the catalytic tyrosines move from their observed trans positions to being in cis, and then the nearby terminal 3' OH groups on the DNA resolve the covalent intermediates. Voila: sealed donor ends and a transposon circle!
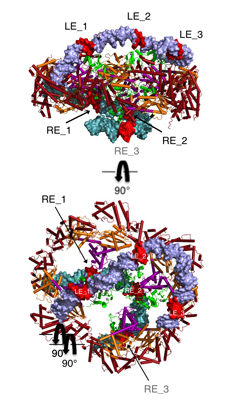
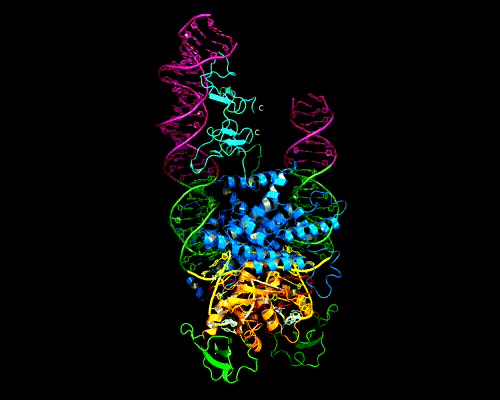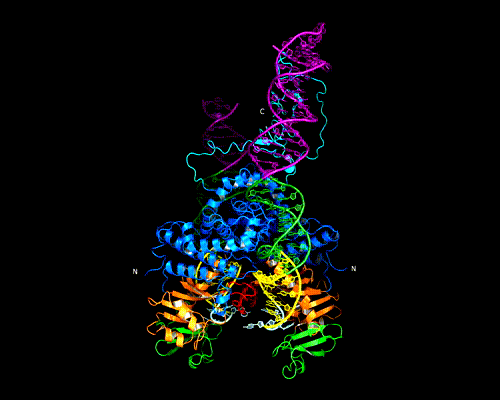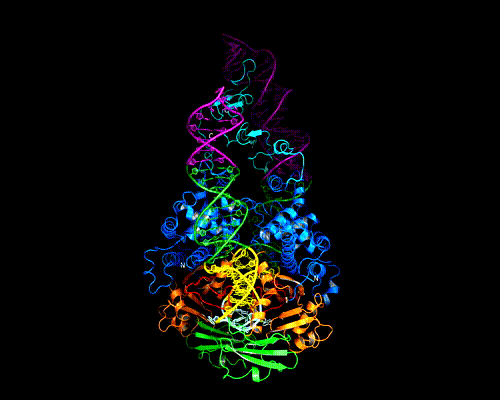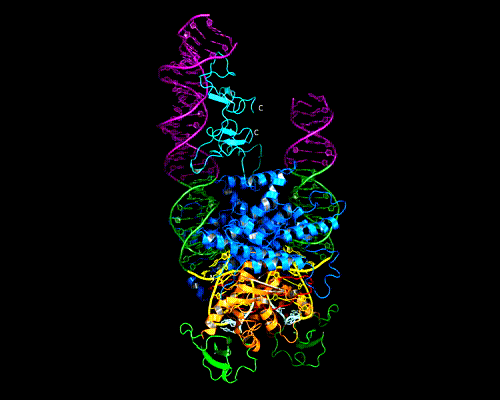
The Hermes Octamer
Hermes assembles as an octameric ring, a very unusual arrangement for a DNA transposase. The octamer is a tetramer of dimers, and biochemical data indicate that a dimer by itself is sufficient to carry out all DNA cutting and joining steps needed for transposition, but only in a test tube at very low ionic strengths. In cells, the octamer must exist for transposition to happen.
As shown in the figure, we think that the octameric ring is needed so that Hermes can make high affinity interactions with its long transposon ends, and that a dimer cannot interact in this way. (The LE_1-3 and RE_1-3 labels indicate the positions of repetitive DNA motifs found within the transposon ends.) A fascinating question is how this assembly assures that both tips of the two transposon ends are bound by the same dimer of the ring. We know this must be the case as only then would it be possible for Hermes to attack the two strands of target DNA eight basepairs apart to achieve integration of the transposon (an absolutely conserved feature of Hermes transposition). Quite clearly, Hermes must employ some trick to achieve this and we hope to discover what this trick is.
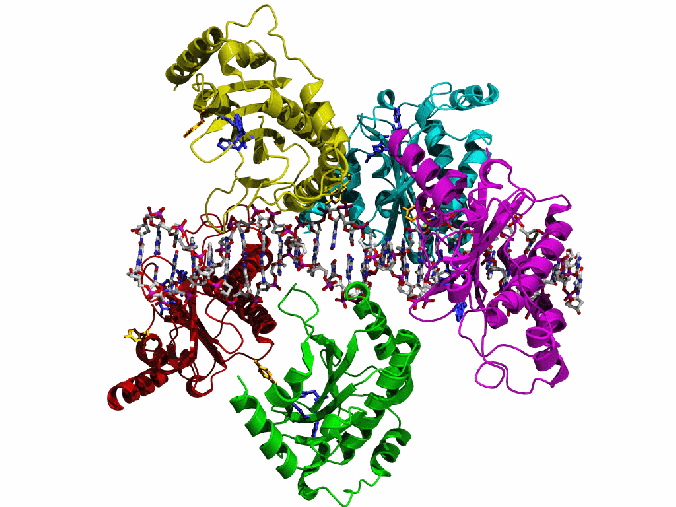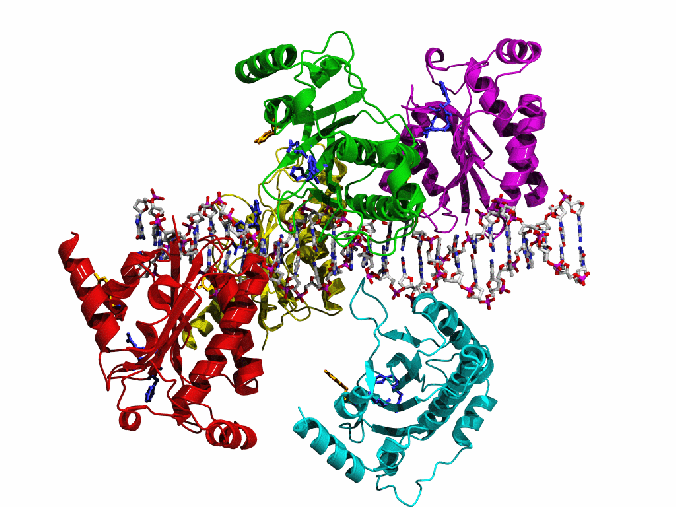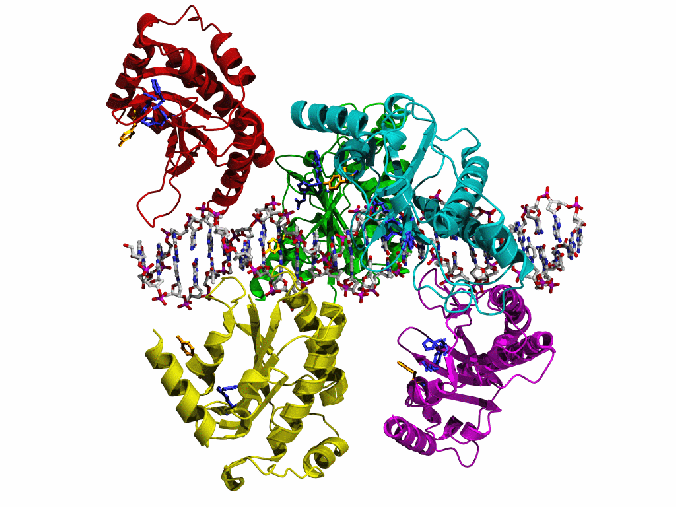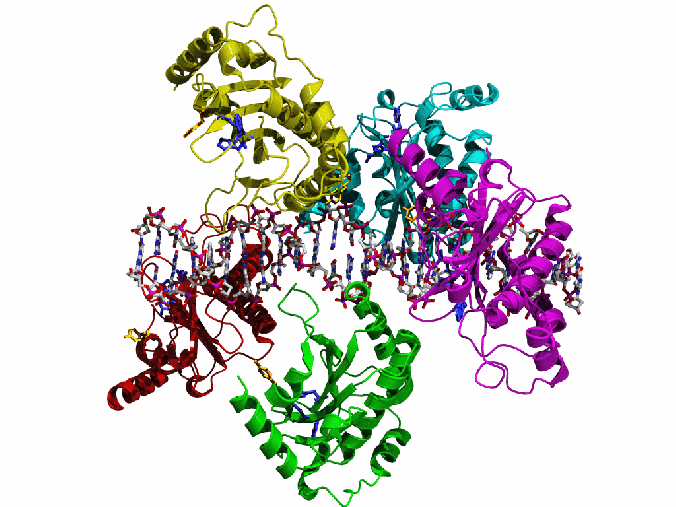
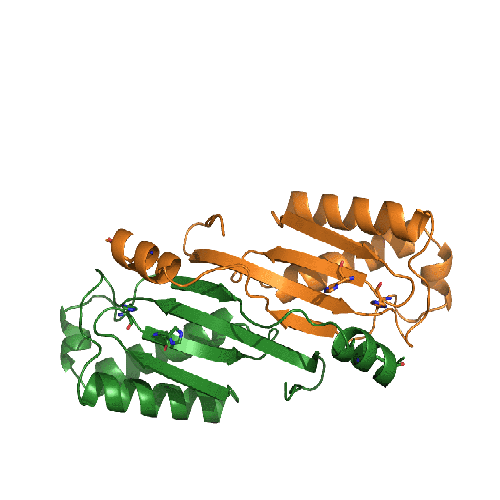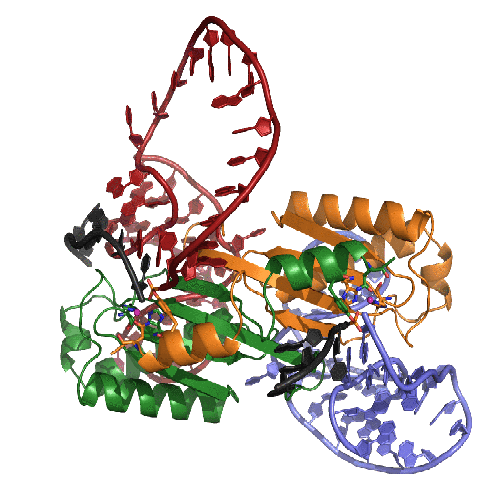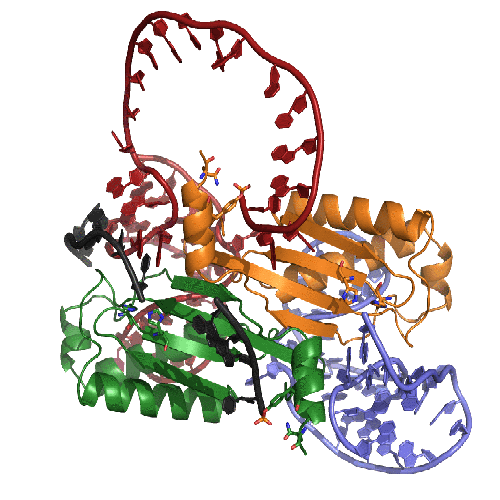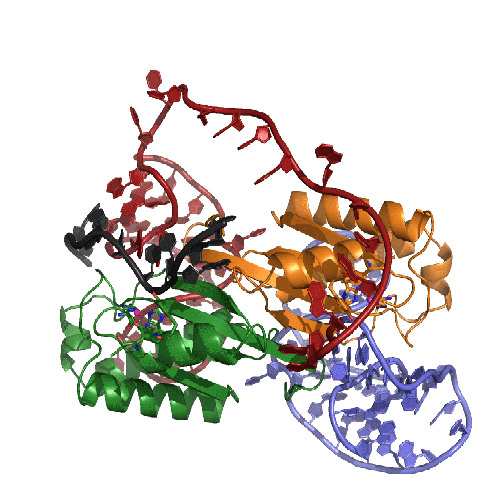
AAV Rep bound to the viral Rep Binding Site (RBS)
This shows how five Rep DNA-binding domains are arranged around the Rep Binding Site that is found in the AAV viral genome. The RBS has several direct repeats of a 5'-GCTC-3' motif. A similar - but not identical - RBS sequence is also found at AAVS1, the locus in human chromosome 19 where AAV integrates.
The structure reveals that the Rep DNA-binding domains spiral around the DNA axis, offset from one another by four base pairs. Two protein structural elements that are important for RBS binding are a surface loop which sits in the major groove and an alpha helix which is inserted into the minor groove. Each Rep monomer interacts with DNA by providing a loop to one tetranucleotide repeat and an alpha helix to the adjacent repeat in a manner reminiscent of tweezers grasping DNA.
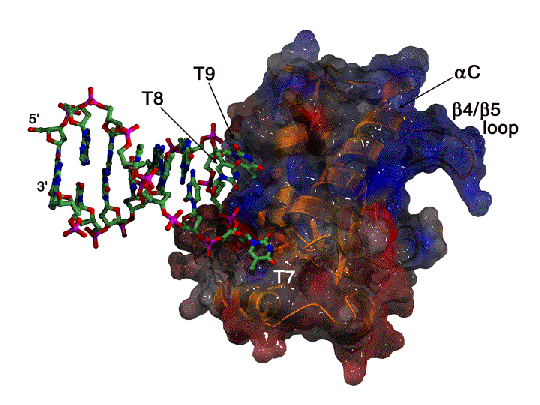
Recognition of an ITR hairpin
The AAV genome is single-stranded, but each end (known as the Inverted Terminal Repeat, or ITR) has palindromic sequences that allow the formation of a three-way DNA junction with two short (9 bp) hairpin arms. We determined that Rep binds specifically to one of these hairpins, and the structure of the complex is shown here.
At the tip of the hairpin, three consecutive bases, T7-T9, interact directly with Rep. T7 is flipped out, while T8 and T9 are in conformations that allow continued helical stacking of the hairpin stem into the loop. The flipped-out base is inserted into a pre-formed surface pocket of Rep, where all of the potential hydrogen bond donors and acceptors in T7 are used.
Why does Rep bind to this part of the viral ITR? We suspect that when Rep is anchored to two parts of the viral DNA - this hairpin and the RBS - this causes a conformational change or a distortion of the DNA. Perhaps this is needed for the other part of Rep, a hexameric helicase domain, to assemble.


Josh Chappie, Alison Hickman, Andrea Voth, Simon Messing, Jenna Maggin and Fred Dyda