The structural biology of Toll-like receptors
Our research focuses on the structural biology of proteins involved in innate immunity. We study the structure of these proteins by X-ray crystallography and a variety of biochemical and biophysical techniques. The membrane-bound Toll-like receptors (TLRs) trigger innate immune responses following recognition of a wide variety of pathogen-derived compounds. Although the ligand-induced dimerization of these receptors has many common features, the nature of the interactions of the TLR extracellular domains with their ligands varies markedly between TLR paralogs.
Botos, I., Segal, D.M., Davies, D.R. The Structural Biology of Toll-Like Receptors. Structure 19: 447-459, 2011. Review.
Wang, Y., Liu, L., Davies, D.R., Segal, D.M. Dimerization of Toll-like Receptor 3 (TLR3) is required for Ligand binding. J. Biol. Chem. 285: 36836-36846, 2010.
Fitzkee, N.C., Masse, J.E., Shen, Y., Davies, D.R., Bax, A. Solution Conformation and Dynamics of the HIV-1 Integrase Core Domain. J. Biol Chem, 285: 18072-18084, 2010.
Li, M., Chen, C.Q., Davies, D.R., Chiu, T.K. An Induced-fit Mechanism for Prolyl Endopeptidase. J.Biol.Chem. 285: 21487-495, 2010.
Botos, I., Liu, L., Wang, Y., Segal, D.M., Davies, D.M. The Toll-like Receptor 3 signaling complex with dsRNA. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1789: 10667-674, September-October 2009. Review. Full text
Liu, L., Botos, I., Wang, Y., Leonard, J.N., Shiloach, J., Segal, D.M., Davies, D.R. Structural basis of Toll-like receptor 3 signaling with double-stranded RNA. Science, 320, 379-381, 2008. Abstract Full text
3CIY
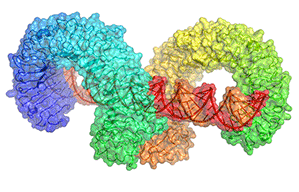
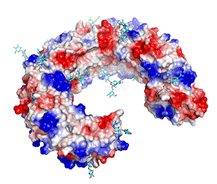
Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
(2008) Science 320: 379
Related structures: 3CIG
2A0Z
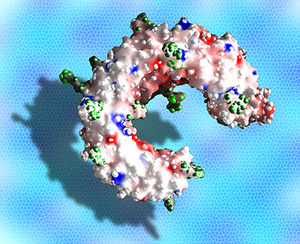
The molecular structure of the Toll-like receptor 3 ligand-binding domain
(2005) Proc.Natl.Acad.Sci.USA 102: 10976
3IVM
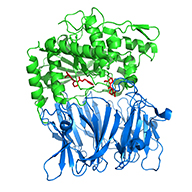
Induced-fit mechanism for prolyl endopeptidase,
(2010) J.Biol.Chem.285: 21487
Related Structures: 3IUR, 3IUQ, 3IUN, 3IUM, 3IUL, 3IUJ
1YRF
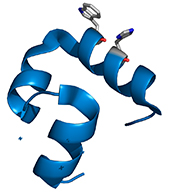
High-resolution x-ray crystal structures of the villin headpiece subdomain, an ultrafast folding protein.
(2005) Proc.Natl.Acad.Sci.Usa 102: 7517
1DHS
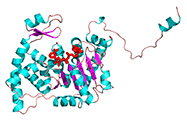
Crystal structure of the NAD complex of human deoxyhypusine synthase: an enzyme with a ball-and-chain mechanism for blocking the active site.
(1998) Structure 6: 23
Related Structures: 1RQD, 1ROZ, RLZ
1HQO

The crystal structure of the nitrogen regulation fragment of the yeast prion protein Ure2p.
(2001) Proc.Natl.Acad.Sci.USA 98: 1459
1TTP
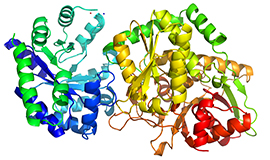
Exchange of K+ or Cs+ for Na+ induces local and long-range changes in the three-dimensional structure of the tryptophan synthase alpha2beta2 complex.
(1996) Biochemistry 35: 4211
Related structures: 1UBS, 1TTQ, 1BKS
1D3R
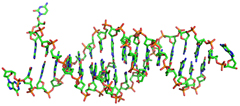
Structure of a triple helical DNA with a triplex-duplex junction.
(1999) Biochemistry38: 16810
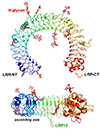
Human TLR3 extracellular domain structure
Despite the wide range of ligands recognized by TLRs, the receptors share a common structural framework: each with an N-terminal ligand recognition domain, a single transmembrane helix, and a C-terminal cytoplasmic signaling domain. The extracellular, ligand recognition domain is formed from leucine-rich repeats, arranged in the form of a horseshoe.
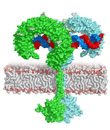
Model of full-length TLR3 bound to dsRNA
Typically, upon ligand binding, two extracellular domains form an "m"-shaped dimer sandwiching the ligand molecule bringing the transmembrane and cytoplasmic domains in close proximity and triggering a downstream signalling cascade.


